Although foot-and-mouth disease virus (FMDV) is among the most economically important livestock diseases in the world, our understanding of its epidemiology in regions where it is endemic is poorly understood. FMDV serotypes A, O, SAT1 and SAT 2 are endemic in Africa with African buffalo known as carriers of SAT serotypes. Epidemiological and genetic evidence is fairly conclusive that buffalo populations are the source of the disease for cattle in the Southern Africa regions. Little is known about the situation in East Africa yet it has amongst the most complex FMDV situations in the world due to high viral diversity, unrestricted livestock movement, and presence of wildlife reservoirs. Although wildlife-livestock contact in East Africa is more frequent and intimate due to shared rangelands, the role of buffalo as an FMDV reservoir has not been resolved due to lack of information on the genetic diversity of FMDV circulating in buffalo.

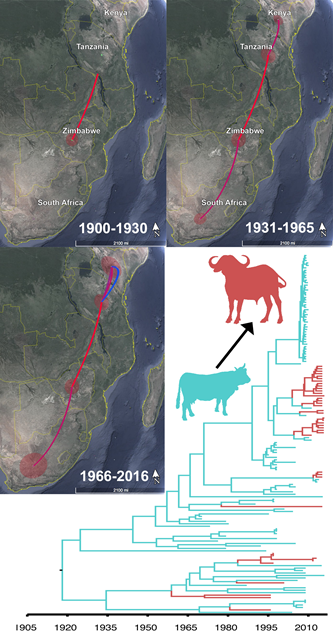
We sequenced 80 buffalo-origin FMD viruses and examined the evolutionary epidemiology of currently circulating clades of SAT1 and SAT2 FMDV in East Africa. Our analyses suggest that currently circulating SAT1 viruses in East Africa originated in Zimbabwe, whereas Kenya is the likely country of origin for contemporary SAT2 viruses.
We also show that cattle are the likely source of the SAT1 and SAT2 clades currently circulating in East Africa, though buffalo may still have an ancestral role deeper in the past. Our results suggest that the majority of SAT1 and SAT2 in cattle comes from other livestock rather than buffalo, with limited evidence that buffalo serve as reservoirs for cattle. Insights from the present study highlight the role of transboundary spread, most likely through cattle movements and other anthropogenic activities, in shaping the evolutionary history of FMDV in East Africa.
Blog by Moh Alkhamis and Kim VanderWaal (Kuwait University & University of Minnesota)
For the full article, see:
Omondi, G, Alkhamis, MA, Obanda, V, et al. Phylogeographical and cross‐species transmission dynamics of SAT1 and SAT2 foot‐and‐mouth disease virus in Eastern Africa. Mol Ecol. 2019; 28: 2903– 2916. https://doi.org/10.1111/mec.15125
