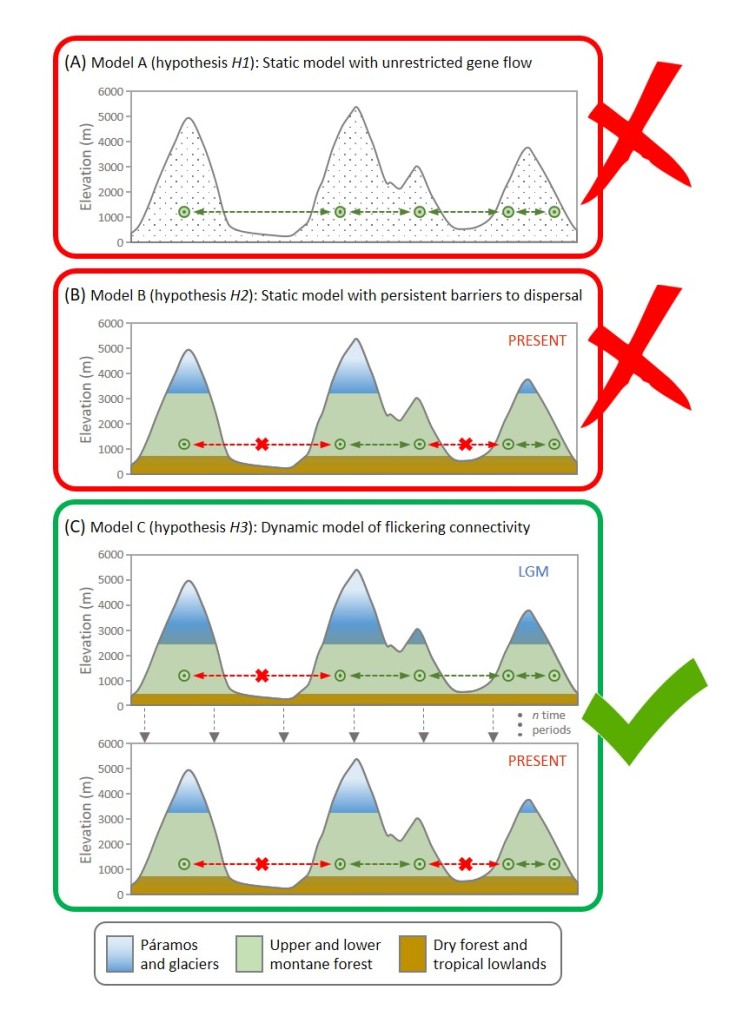
In a recent paper in Molecular Ecology, Ortego et al. integrated genomic data of the Andean oak (Quercus humboldtii) from the Colombian Cordilleras and spatiotemporally explicit inference frameworks to test competing hypotheses about the demographic trajectories of the species. Their analyses support a dynamic model of flickering population connectivity since the Last Glacial Maximum, emphasizing the interplay between spatial configuration of geographical barriers to dispersal and Quaternary climatic oscillations in shaping demographic trajectories of montane oak forests, and further, tropical montane biotas.
We sent a number of questions to lead author of this work, Joaquín Ortego, to get more detail on this study.

What led to your interest in this topic / what was the motivation for this study?
Relatively little is known about the impacts of Pleistocene glacial-interglacial cycles on tropical biotas and the different models proposed to explain their hypothetical demographic and distributional shifts in response to past climate changes have seldom been formally tested using genomic data. Thus, the main motivation of our study was to shed some light into the processes that have determined the demographic dynamics of Andean montane forests, with a particular emphasis on the potential role of Quaternary climatic oscillations on shaping population connectivity and isolation. To set up our hypotheses, we found very inspiring the extensive palaeoecological research performed in the tropical Andes during the past decades (e.g., Hooghiemstra & Van der Hammen, 2004) and the alternative distributional and demographic models proposed in previous studies (Ramírez-Barahona & Eguiarte, 2013; Flantua et al., 2019). By integrating genomic data, species distribution modelling and demographic simulations we could explicitly test hypotheses that have remained largely unanswered for a long time.
What difficulties did you run into along the way?
From an analytical point of view, the main difficulty we found was creating the different spatially explicit scenarios we aimed to test, particularly the one incorporating reconstructions of past species distribution at a fine temporal resolution. It would have been also nice to analyze populations across the entire distribution of the species, but it was not possible due to logistic limitations and time-constrains during our fieldwork.
What is the biggest or most surprising innovation highlighted in this study?
The most interesting aspect of our study is that we demonstrate that the Andean montane oak forest is not a static ecosystem. On the contrary, our genomic data and demographic modelling indicate that Andean montane forests have experienced changes in population connectivity and isolation through time linked to Quaternary climatic oscillations. It is also surprising the high genetic fragmentation of tropical oak populations, which has likely resulted in most of them have experienced idiosyncratic demographic trajectories. A totally unexpected result we obtained – actually inspired by the comments of one of the Reviewers – was that genetic differentiation was best explained by elevational dissimilarity among populations (i.e., “isolation-by-elevation”) than by the geographical distance that separate them (i.e., “isolation-by-distance”), pointing to local adaptation processes linked to the contrasting environmental conditions prevailing at different elevational ranges. This result is pretty cool in my opinion, as such a clear effect of elevation dissimilarity on patterns of gene flow among populations has been very rarely reported in the literature. Definitively, I would like to explore this aspect more in depth in the future, analyzing more populations and testing for genomic signatures of local adaptation or phenological mismatching linked to environmental dissimilarity.
Moving forward, what are the next steps in this area of research?
From my perspective, the next natural step would be to extend our analyses to multiple species of the different vegetation belts (i.e., lowlands, lower montane forest, upper montane forest, and paramos) within a community-level comparative framework. This would allow testing for concordant vs. species-specific demographic trajectories across multiple taxa, which might help the reach more general conclusions about the role of past climatic oscillations on the dynamics of tropical ecosystems (e.g., Helmstetter et al., 2020; Prates et al., 2016). The aforementioned testing of “isolation-by-elevation” in other taxa might also provide interesting insights into the proximate ecological and evolutionary processes that have contributed to geographical diversification in the tropical Andes and the extraordinary levels of local endemism characterizing this biodiversity hotspot.

What would your message be for students about to start developing or using novel techniques in Molecular Ecology?
My message would be that they do not get lost in the massive genomic data that we can now easily obtain and focus on the questions and hypotheses that they want to address. Curiosity moves science forward, simple observation and exploration is the beginning of everything!
What have you learned about methods and resource development over the course of this project?
The 100-year resolution bioclimatic layers now available through the CHELSA-TraCE21k database (https://chelsa-climate.org/chelsa-trace21k/; Karger et al., 2023) allowed us to get a more realistic picture of the distributional dynamics of the focal species, which was key for our demographic inferences and interpreting the data (e.g., changes in elevational displacements through time). I think this new resource will be very useful for future biogeographic and distributional studies.
Describe the significance of this research for the general scientific community in one sentence.
Tropical ecosystems were not spared from the impacts of Pleistocene climatic changes.
Describe the significance of this research for your scientific community in one sentence.
Andean montane forest follow a model of flickering genetic connectivity linked to Quaternary climatic oscillations.
References
Flantua, S. G. A., O’Dea, A., Onstein, R. E., Giraldo, C., & Hooghiemstra, H. (2019). The flickering connectivity system of the north Andean paramos. Journal of Biogeography, 46(8), 1808–1825. https://doi.org/10.1111/jbi.13607
Helmstetter, A. J., Bethune, K., Kamdem, N. G., Sonke, B., & Couvreur, T. L. P. (2020). Individualistic evolutionary responses of central African rain forest plants to Pleistocene climatic fluctuations. Proceedings of the National Academy of Sciences of the United States of America, 117(51), 32509–32518. https://doi.org/10.1073/pnas.2001018117
Hooghiemstra, H., & Van der Hammen, T. (2004). Quaternary ice-age dynamics in the Colombian Andes: Developing an understanding of our legacy. Philosophical Transactions of the Royal Society of London Series B-Biological Sciences, 359(1442), 173–180. https://doi.org/10.1098/rstb.2003.1420
Karger, D. N., Nobis, M. P., Normand, S., Graham, C. H., & Zimmermann, N. E. (2023). CHELSA-TraCE21k – High-resolution (1 km) downscaled transient temperature and precipitation data since the last glacial maximum. Climate of the Past, 19(2), 439–456. https://doi.org/10.5194/cp-19-439-2023
Prates, I., Xue, A. T., Brown, J. L., Alvarado-Serrano, D. F., Rodrigues, M. T., Hickerson, M. J., & Carnaval, A. C. (2016). Inferring responses to climate dynamics from historical demography in neotropical forest lizards. Proceedings of the National Academy of Sciences of the United States of America, 113(29), 7978–7985. https://doi.org/10.1073/pnas.1601063113
Ramírez-Barahona, S., & Eguiarte, L. E. (2013). The role of glacial cycles in promoting genetic diversity in the neotropics: The case of cloud forests during the last glacial maximum. Ecology and Evolution, 3(3), 725–738. https://doi.org/10.1002/ece3.483
Featured study
Ortego, J., Espelta, J. M., Armenteras, D., Díez, M. C., Muñoz, A., and Bonal, R. (2023) Demographic and spatially-explicit landscape genomic analyses in a tropical oak reveal the impacts of late Quaternary climate change on Andean montane forests. Molecular Ecology, 32(12), 3182-3199. https://doi.org/10.1111/mec.16356
